Great Expectations
Great Expectations
Great Expectations is a framework that enables you to create and maintain a set of expectations for what your data should look like and contain. Specifically the project indicates it is aimed at: “helping data teams eliminate pipeline debt, through data testing, documentation, and profiling.”
In our desire to have more automated Data Science development processes there are a few areas that should be considered:
During build/deployment:
- Automated deployments - CI/CD pipelines to create and test project environment and dependencies
- Automated unit testing - typically used during build/deployment to make sure that logical functionality is correct
During continuous learning & inference:
- Automated model validation - used after model training to assess the accuracy of a built model
- Data drift monitoring & detection - used to make sure that inference data matches training data characteristcs (new columns, new distributions, new data types, etc.)
Great Expectations helps with this last point allowing the integration of “data checks” within established Data Science pipelines. We can picture a Data Science DAG as possibly looking like the below where data validation rules are created during a training cycle and then reapplied during an inference cycle:
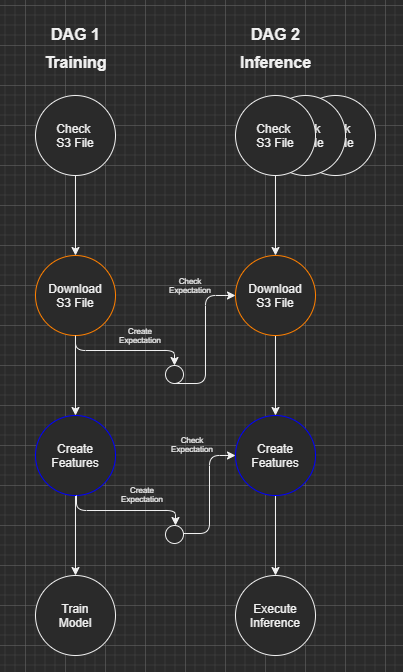
Visual created using the excellent DrawIO VSCode plugin.
Example Project
In order to explore Great Expectations more fully we will setup an example tutorial of how usage would likely look in real-world situations. This walk-through will involve the followign steps:
- Download an initial training set
- Execute Exploratory Data Analysis (EDA) on data-set
- Initialize a Great Expectations suite
- Create initial expectations automatically using basic profiling
- Modify created expecations to more closely match data intentions
- Build a Machine Learning pipeline that leverages the created Expectations to confirm data quality
- Download a new data-set for inference using the built model including Expectation validation
- Confirm that validation prevents inference and review the resulting errors
Code repository can be found here. File used below are wine_quality_1.parquet and wine_quality_2.parquet.
TL;DR
The training sets will be built using the UCI Wine Quality data-set. The initial training set will be based only on red wines whereas the inference will attempt to execute on both red and white wines. Great Expectations will notice difference in indicated type (categorical variable) as well as numerical distribution changes preventing predictions on the inference data-set.
Setup
As usual start by creating a virtual environment and installing the required dependency great-expectations. A reference Pipfile can be found here. Side Note: pip install great-expectations is a fantastic Computer Science meets Literature joke.
You can review wine_quality_1.parquet to understand data structure and basic characteristics:
df = pd.read_parquet ("../data/sample/wine_quality_1.parquet")
df.describe(include="all")
View notebook to see expected output.
Initialize Great Expectations
Initialize a great expectations project via great_expectations init. This will create a standard project structure in great_expectations folder that will contain expectation configurations as well as validation run results.
In this case most of the defaults for initialization are ok:
- “Y” to create a standard directory
- “Y” to create a DataSource
- “1” to use Local Files
- “1” to use Pandas
- “data” to set data source path to a root of ./data
- “data_ds” to set the name for the created DataSource
- “N” to prevent profiling of data
The above results in the creation of the great_expectations project folder including a yaml defined DataSource which sets up a general local, Pandas file reader. For more complicated DataSource like databases or remote storage this pre-configured interface would potentially have more value.
data_ds:
data_asset_type:
class_name: PandasDataset
module_name: great_expectations.dataset
batch_kwargs_generators:
subdir_reader:
class_name: SubdirReaderBatchKwargsGenerator
base_directory: ..\data
class_name: PandasDatasource
module_name: great_expectations.datasource
Creating Data Expectations
To automatically create a suite of expectations you can use great_expectations suite scaffold data_ds.
At the prompts:
- “1” to list data assets
- “1” to select the available data (the CLI will automatically find the wine_quality_1.parquet file)
Great Expectations uses Jupyter Notebooks as a front-end for configuration. Many of the great_expectation CLI commands result in the launch of the Jupyter server and a browser view to execute or edit Notebook fields.
Jupyter Notebooks will open to provide default settings for data profling. In this case we would like to profile all columns except for quality since quality will be our predicted target variable.
included_columns = [
'fixed acidity',
'volatile acidity',
'citric acid',
'residual sugar',
'chlorides',
'free sulfur dioxide',
'total sulfur dioxide',
'density',
'pH',
'sulphates',
'alcohol',
# 'quality',
'type'
]
Once you have edited and executed the Jupyter cells the expectations suite will be created at great_expectations/expectations/data_ds.json. The assocated data docs will be opened.
At this point you will notice that the data docs will indicate that the validation suite is failing. In particular as the profiler uses a sample of data to create initial expectations it can produce expectations that are not always well suited for your data. You can create your own custom auto-profiling logic if desired.
Use great_expectations suite edit data_ds to open up Jupyter Notebooks to directly edit the auto-generated expectations. In particular you would want to change:
- Remove fixed expectations on table row count
- Remove “unique value” constraints on all columns
- Add a check for colume “type” that it must be “red” (since that is the only value currently used in column)
Automatically generated expectations should never be used directly in production. You would still want Data Science and Data Management to review and modify created expectations to make sure that data is being correctly qualified.
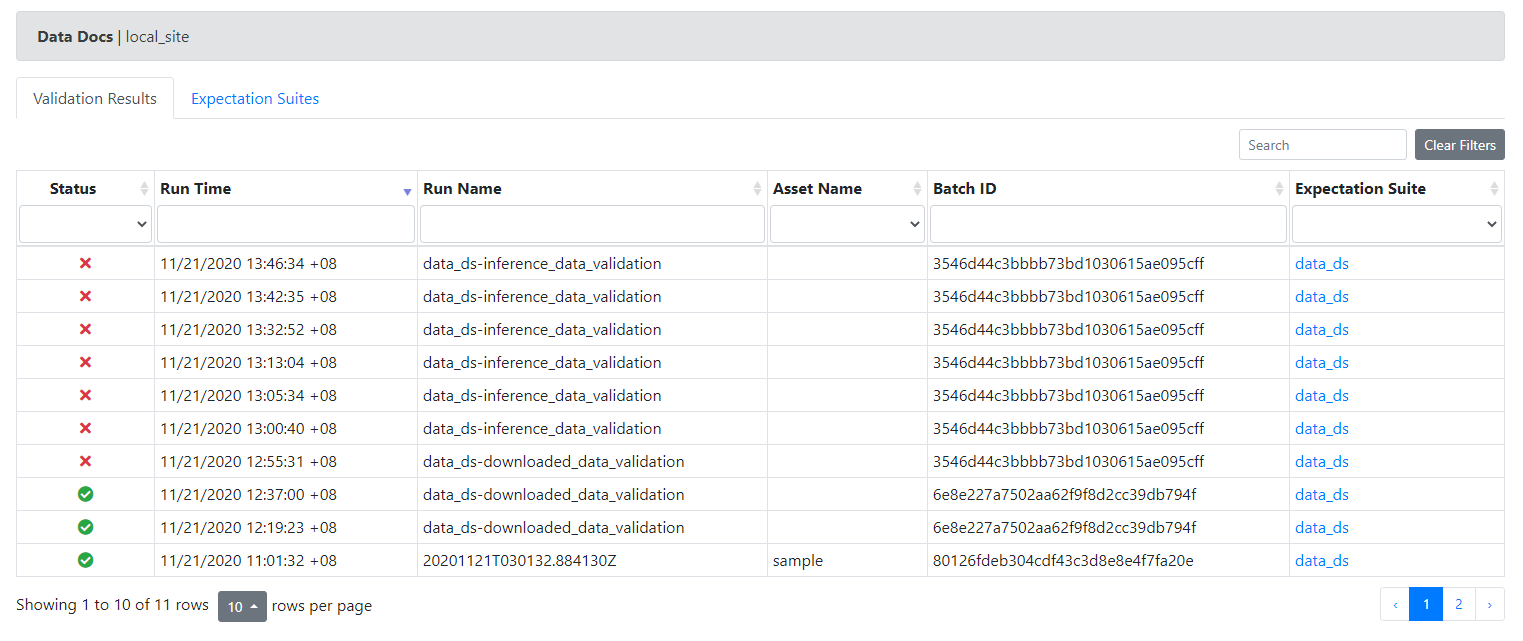
When you save and re-run your validation you should now see that your data-docs indicate a successful run. You can review historical validation runs at /great_expectations/uncommitted/data_docs/local_site/index.html. Clicking on results will take you to summary of failures.
Integrating into pipeline
For this example I am using the Luigi pipeline framework but in general the below would be pretty consistent in other frameworks like Airflow.
Create a GreatExpectationsValidationTask that can be inserted into a pipeline to execute a validation step:
Full implementation can be viewed in repository but key areas of interest include:
# Get a Great Expectations context:
context = ge.data_context.DataContext()
# Create a batch to validate vs. a local file referenced via DataSource:
batch_kwargs = {"path": data_file_path, "datasource": self.datasource_name}
batch = context.get_batch(batch_kwargs, self.datasource_name)
# Create a run identifier that is meaningfully related to the pipeline execution:
run_id = {
"run_name": self.datasource_name
+ "-"
+ self.validation_sub_dir
+ "_validation", # insert your own run_name here
"run_time": datetime.datetime.now(datetime.timezone.utc),
}
# Validate data batch vs. expectation suite. Using run_validation_operator
# instead of batch.validate() to invoke data docs update operations.
results = context.run_validation_operator(
"action_list_operator", assets_to_validate=[batch], run_id=run_id
)
This will take whatever files are passed into the step and will validate them vs. the current expectations associated with the provided datasource. This generalized task can now be inserted into both training and inference pipelines to insure consistent data across both pipeline executions (which is what we are interested in to make sure that our prediction accuracy is not eroded via data drift).
Pipeline Execution
Implementations for the two tasks can be found in repository to execute both training and inference via XGBoost:
Implemented code is expecting to read from S3 bucket as data-source. You can either change data implementation to point to your own S3 bucket or change data implementation to read from locally downloaded files.
Running the training DAG using the wine_quality_1.parquet file results in no errors as expected. However running the inference DAG using wine_quality_2.parquet results in an error that prevents pipeline from completion and outputs the following errors:
Failed: (fixed acidity) expect_column_max_to_be_between
Failed: (fixed acidity) expect_column_mean_to_be_between
Failed: (fixed acidity) expect_column_median_to_be_between
Failed: (fixed acidity) expect_column_quantile_values_to_be_between
Failed: (residual sugar) expect_column_max_to_be_between
Failed: (residual sugar) expect_column_mean_to_be_between
Failed: (residual sugar) expect_column_quantile_values_to_be_between
Failed: (free sulfur dioxide) expect_column_max_to_be_between
Failed: (free sulfur dioxide) expect_column_mean_to_be_between
Failed: (free sulfur dioxide) expect_column_median_to_be_between
Failed: (free sulfur dioxide) expect_column_quantile_values_to_be_between
Failed: (total sulfur dioxide) expect_column_max_to_be_between
Failed: (total sulfur dioxide) expect_column_mean_to_be_between
Failed: (total sulfur dioxide) expect_column_median_to_be_between
Failed: (total sulfur dioxide) expect_column_quantile_values_to_be_between
Failed: (alcohol) expect_column_max_to_be_between
Failed: (type) expect_column_distinct_values_to_be_in_set
Errors can be explored in more detail using the generated data_docs.
In this case our Expectation suite is helpfully identifying that means and medians of columns have substantially shifted and that a new type of value is now present in “type” columns. At this point we would need to assess if:
- We update our expectations and allow existing model to run
- We rebuild our model with the new data-set and set new expectations
Additional automation opportunity
In the above example implementation an initial training data-set is manually validated through the use of the Exploratory Data Analytics. This results in the analyst being responsible for the creation of the resulting expectations. This is fine (and maybe even preferable) in the case where training is infrequent, but if data drift is expected and training is frequent this could pose a bottleneck. It would be possible to expand on the Great Expectations auto-scaffolding to create a consistent set of automatically generated expectations for any training data-set. This auto-generated validation could then be used by inference to have a fully automated data quality assesment that enforced expectations between learning and inference.
In the case of manual expectation creation one could ask why bother applying validation to training task? This can be considered optional but it does provide a nice check to make sure that your expectations are well formed and reflective of training data. This can help catch any accidentally overly restrictive expectations - e.g. if the validation step fails for training cycle then you have formed an incorrect expectation.
Benefits
In this particular example it would be a bad idea to use the quality prediction model based on red wine to predict on white wine so as good Data Scientists we would look to rebuild the model and re-deploy. If this was a priority business decision being recommended it would critical that we had indication that predicted outcomes were likley to be incorrect BEFORE we act on those predictions (as opposed to learning after the fact). Great Expectations helps to pre-emptively safe-guard us against issues with data-discrepencies and drift and is something that should be considered as a needed part of every ML Pipeline.
